Genetic Modification Techniques and Applications
Gene modification, editing or engineering, do you know the difference, the routes to success or how they benefit us?
Complete the form below to unlock access to ALL audio articles.
Genetic modification previously hit the headlines in relation to “Frankenstein crops” and received bad press as an “unnatural” practice. However, mankind has been manipulating genetics for eons, initially through selective breeding and more recently through the use of techniques that speed up this process in very controlled and precise ways.
What is gene editing?
Gene editing vs genetic engineering
Creating a gene knockout, deletion mutation, insertion mutation, substitution mutation or point mutation
Nonhomologous end joining (NHEJ) – ligation without matching sequences
Homology directed repair (HDR) - homologous recombination repair
Genetic modification techniques
In this article, we will consider genetic modification strategies, including genome editing and genetic engineering, the techniques used to achieve them and purposes to which they are being applied.
What is gene editing?
Gene editing is a form of genetic modification and is the process by which the DNA sequence of an organism is modified or deleted. This is normally carried out to obtain desirable effects, such as the modification of a protein, to produce a preferable phenotype or to prevent a problematic gene from being transcribed. Gene editing has been described as being a bit like editing a document. Scientists can find a specific “word” or “phrase”, in this case a DNA sequence or gene, and then delete it, change it for another “word” or add additional “words” to improve it.
Gene editing vs genetic engineering
Like gene editing, genetic engineering is a form of genetic modification. However, as well as including the modification or deletion of a base or region of an organism’s genetic code, genetic engineering also includes the temporary or permanent insertion of foreign DNA from another organism, sometimes called a transgene. This mix of DNA from multiple sources, the sequences of which are otherwise unlikely to be found in nature, is referred to as recombinant DNA. For the creation, insertion and replication of these sequences in the target host, molecular cloning techniques are key. It should, however, be noted that genome “modification”, “engineering” and “editing” are often incorrectly used interchangeably in literature and discussion.
The distinction between gene editing versus engineering is one that is recognized by numerous regulatory authorities. In many countries, products bound for the food market that contain foreign DNA (genetically engineered) have to go through a rigorous process before reaching the consumer. Those that have been gene edited and contain no foreign DNA have to go through the same process in some countries, whereas in others, they are considered to be possible products of conventional breeding and therefore are able to reach consumers more easily.
Creating a gene knockout, deletion mutation, insertion mutation, substitution mutation or point mutation
As mentioned, different types of genetic modification can be made, including:
- Gene knockout – the removal or inactivation of a target gene.
- Deletion mutation – the removal of one or more nucleotides from a genetic sequence.
- Insertion mutation – the addition of one or more nucleotides into a genetic sequence.
- Substitution mutation – the switching out of one or more nucleotides in a genetic sequence for alternatives.
- Point mutation – the insertion, deletion or substitution of a single base pair in a genetic sequence.
- Gene knock-in – the substitution and/or addition of one or more bases.
Where a mutation introduces a premature stop, truncating the encoded protein, this is called a nonsense mutation.
Where the number of bases added or removed are not a multiple of three, the triplet reading frame for the transcription of DNA to amino acids is disrupted, altering the subsequent amino acid sequence. This is called a frameshift mutation.
Both frameshift and nonsense mutations can result in gene knockout, although the exact consequences will depend on where in the protein this occurs. For example, a single base deletion (in this case naturally occurring) in the fne gene of the bacterium Streptococcus equi, which encodes a fibronectin-binding protein, results in the expression of a binding protein that is still functional but is secreted instead of surface anchored, as the membrane anchor region is no longer produced.1
When it comes to making any genome modification, the DNA of the target organism must be broken in some way for the alteration to be made and then repaired. But to make use of these mechanisms, it is important to understand how DNA repair may occur. Cells have two main repair pathways - nonhomologous end joining (NHEJ) and homology directed repair (HDR).
Nonhomologous end joining (NHEJ) – ligation without matching sequences
NHEJ repairs DNA double-strand breaks (DSBs) and in mammals is the main method of repair for this type of damage throughout the cell cycle, even in non-dividing cells. While some bacterial species are able to perform a form of NHEJ, others, including the laboratory workhorse Escherichia coli (E. coli), lack this pathway and so rely on HDR for the repair of DSBs.
In NHEJ, enzymes capture the broken ends of the DNA, the ends are modified and ligated together irrespective of sequence homology and therefore can be intrinsically mutagenic (Figure 1). A synaptic complex, consisting of two DNA ends, a Ku protein heterodimer and two DNA-dependent protein kinase catalytic subunits (DNA-PKcs) forms, and any non-compatible DNA ends are processed to form ligatable ends. These may be sticky or blunt ends (Figure 2). With sticky ends, one DNA strand has a few bases more than the other and can be ligated to a complimentary sequence on a sticky end at the other side of the break. With blunt ends, there is no overhang that requires a match. This processing step is where mutations tend to be accidentally introduced and the insertion or deletion of a few bases (indel errors) can lead to frameshifts. A ligase complex then repairs the break.
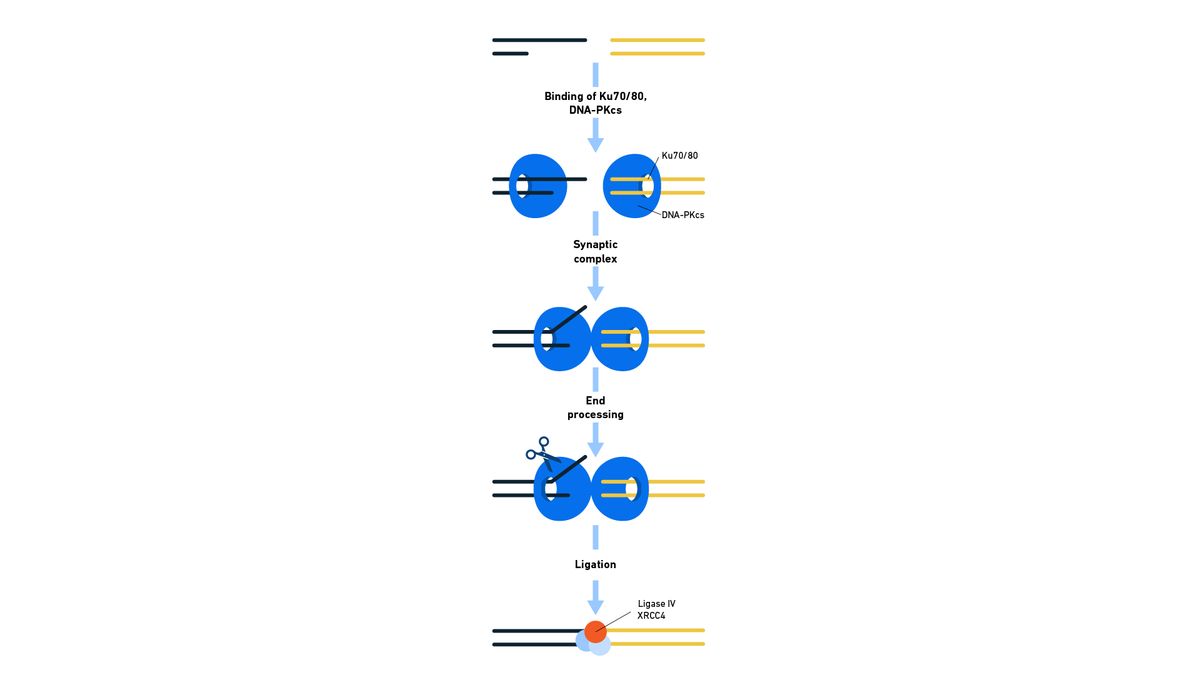
Figure 1: Representation of the steps of NHEJ in eukaryotes, indicating the roles of the enzymes involved. Credit: Technology Networks.
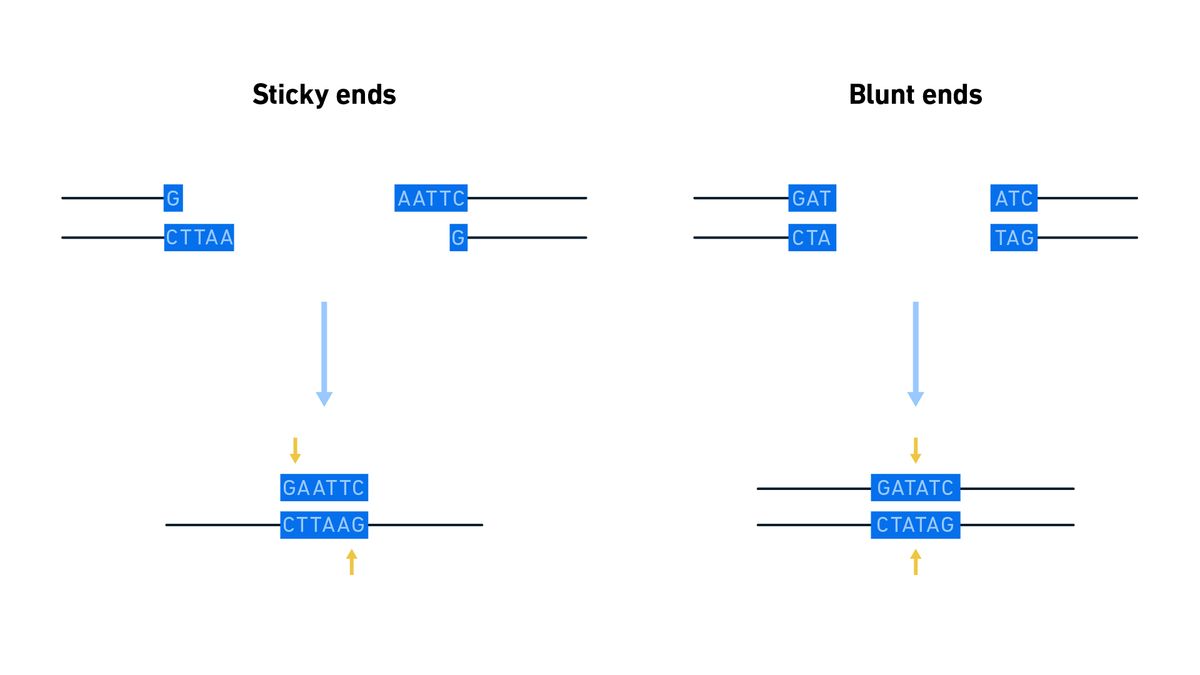
Figure 2: Diagram illustrating sticky (left) versus blunt (right) ends. With sticky ends, the overhanging sequences must be complimentary for ligation to be possible. Credit: Technology Networks.
Unlike eukaryotes, bacterial NHEJ systems are restricted to just two proteins, a Ku homodimer and LigD, a multifunctional ligase, polymerase and nuclease.
Homology directed repair (HDR) - homologous recombination repair
In HDR, the cell uses a homologous, undamaged DNA template to repair the DSB, most commonly via homologous recombination.2 This homologous template may come from sister chromatids in the case of eukaryotes, a sister chromosome from chromosomal replication in the case of bacteria or from a template that has been experimentally added to the cell in the case of genome modification techniques. As HDR uses a template, the natural process is far less error prone than NHEJ.
There are a number of variations on how the process of homologous recombination progresses, but all have a number of core steps in common.
- Nucleolytic processing – following a DSB, the 5′ end of the break is resected to create overhangs – sticky ends.
- Strand invasion – these overhangs provide a substrate for proteins necessary to the process and allow the broken ends to invade and displace one strand of the homologous template sequence.
- DNA synthesis – the undamaged strand is used as a template for repair, forming a structure called a displacement loop (D loop), which transforms to a cross-shaped structure called a Holliday junction.
- Resolution – the recombination intermediates are then resolved and the repair is completed.
In eukaryotes, following DNA synthesis, there are multiple pathways that the repair may take, most commonly, double-strand break repair (DSBR) or synthesis-dependent strand annealing (SDSA) (Figure 3). In DSBR, both 3′ ends form Holliday junctions during strand invasion, facilitating a crossover event, which is important in meiosis, depending on where the products are nicked by endonucleases during resolution. In SDSA, on the other hand, only one 3′ end forms a junction, so no crossover products are produced during resolution, which is important for repair during mitosis. Alternative pathways to homologous recombination include the single-strand annealing (SSA) pathway, which repairs double-strand breaks between two repeat sequences, and the break-induced replication (BIR) pathway, which repairs breaks at replication forks.
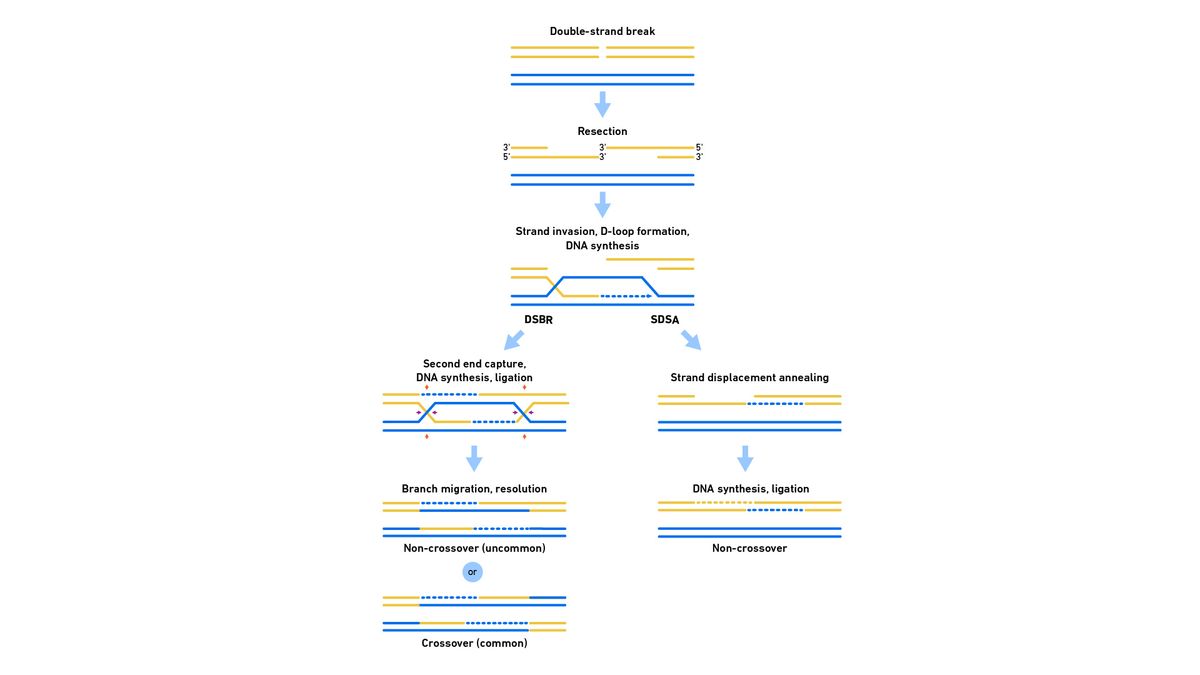
Figure 3: Representation of the steps of HDR in eukaryotes. Credit: Technology Networks.
In bacteria, a similar process occurs with a single Holliday junction to produce either original or recombined copies (Figure 4).
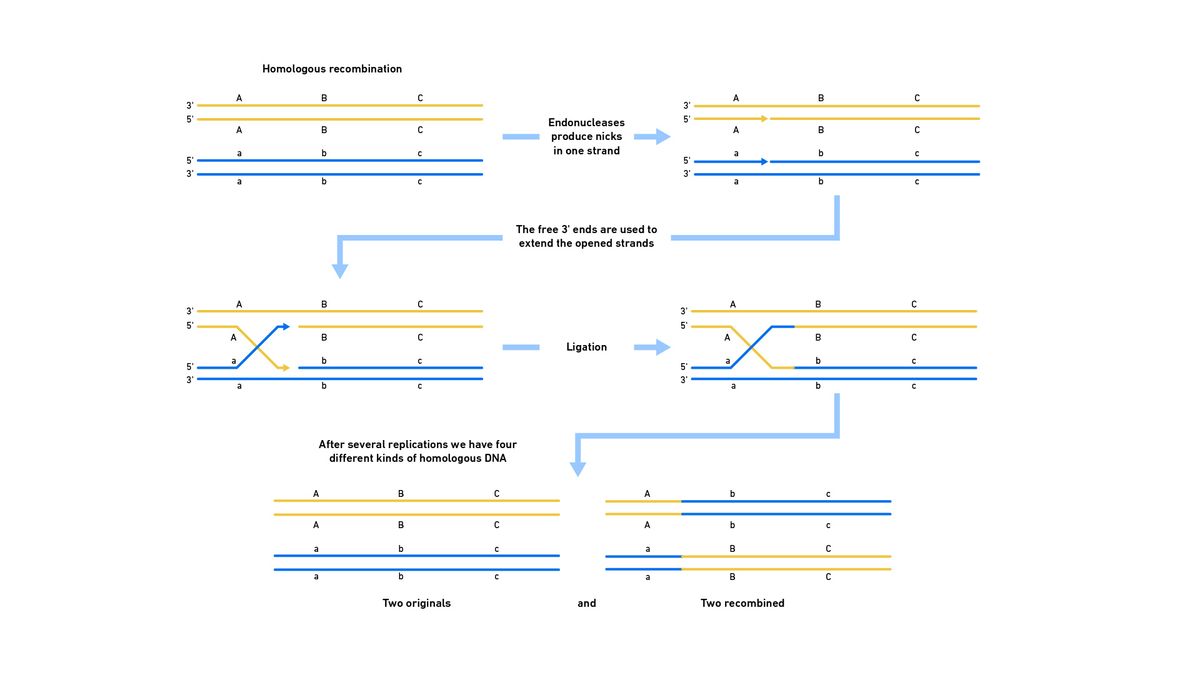
Figure 4: Representation of the steps of HDR in bacteria. Credit: Technology Networks.
While the accuracy of HDR over NHEJ is advantageous, in order to guide repair, HDR requires a homologous sequence or, in the case of genetic engineering, a template with regions of homology flanking the region to be altered. HDR is also slower than NHEJ and, unlike NHEJ, only really works in proliferating cells, as extensive DNA synthesis is required.
However, some bacteria only use HDR, and HDR is the dominant repair mechanism in budding yeast (Saccharomyces cerevisiae) in lab conditions; therefore, understanding which processes can be harnessed in your target species of interest is an important first step in any genome modification work.
In organisms that have capabilities for NHEJ and HDR, aspects such as the cell cycle stage, concentration of donor template, length of the homologous region of a donor template and other factors that affect the activity of each repair system will all contribute to determining which pathway fixes a DSB.
Genetic modification techniques
There are a number of techniques that have been developed to allow scientists to modify genetic sequences specifically, summarized in Figure 5. Whichever technique is used, scientists must first choose the gene or location they wish to modify and the modification they wish to make (insertion, deletion or substitution). They must consider the knock-on impact of any changes made on surrounding genes and ensure any necessary promoter or terminator sequences are present or included. If an insertion or substitution is required, the necessary sequence fragment must be prepared so that it can be delivered by an appropriate means to the cells and act as a template during HDR. Small mutations and base substitutions can be introduced to templates using a process called site-directed mutagenesis, for which the incorporation of the necessary changes into PCR primers used during template synthesis is a popular approach. Let’s consider some of the most common techniques in more detail.
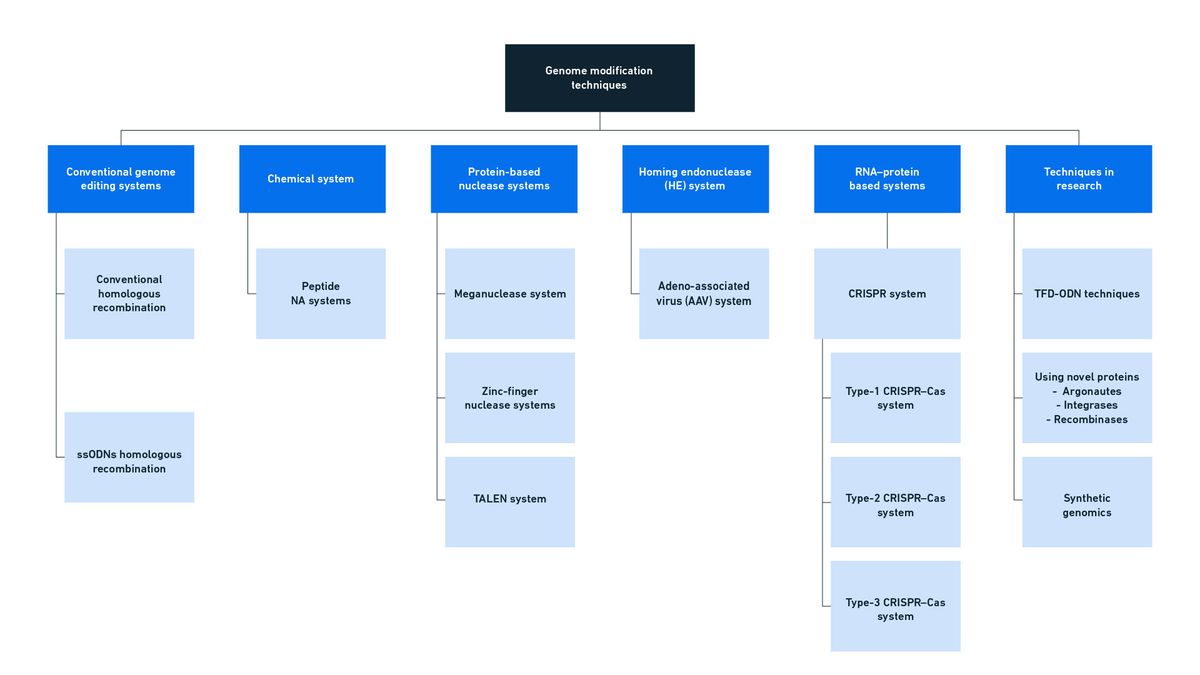
Figure 5: An overview of genome modification techniques. Credit: Technology Networks.
- Gene targeting using homologous recombination
Sometimes referred to as the “conventional” genome modification technique, allelic replacement or gene targeting, uses modified DNA templates created with the help of PCR and restriction enzymes that can be induced to insert into the genome using a cell’s natural homologous recombination DNA repair mechanism. Initially developed in the 1970s and 80s,3 this technique can be used to make a wide range of modifications, from mutating a single base to inserting or deleting a whole gene.
An example of this process in bacteria is summarized in Figure 6. In the example, the aim is to insert a DNA sequence into a target genome. Regions flanking the intended target site are amplified and fused either side of the DNA sequence to be added. This insert is then inserted into a plasmid, which is introduced into the target cells, a method often referred to as molecular cloning. Electroporation is one such technique used to introduce plasmids into cells in a process known as transformation in bacteria and transfection in eukaryotic cells. Other methods to deliver the plasmid include the use of viral vectors. Cells containing the plasmid can be selected for using antibiotic sensitivity specific to the plasmid used. Integration of the insert into the genome can then be stimulated, for example, using a temperature sensitive system. When selective pressure is removed, the plasmid will excise from the genome again, referred to as “curing”, either leaving the cell wild type once more or instead with the new insert which may be classed as “upstream” or “downstream”, depending on which homologous region integration has occurred at. The final purpose of making the mutant will determine if it matters which way round this has occurred or not.
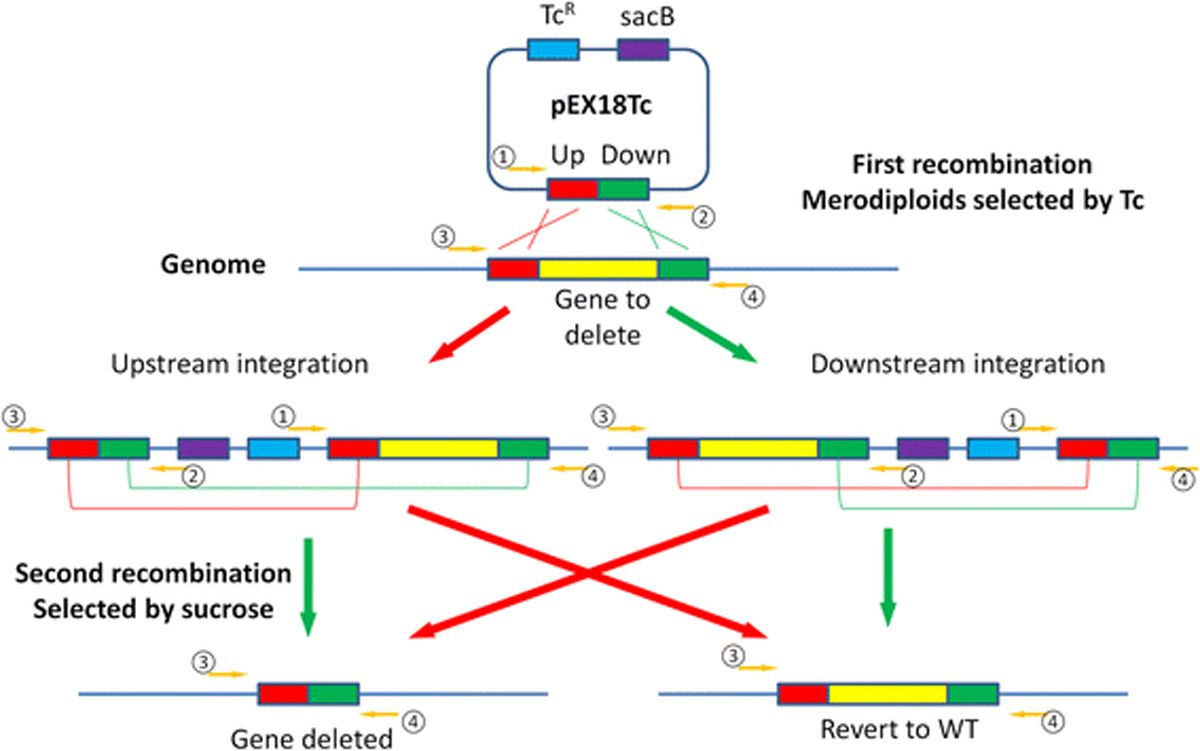
Figure 6: Example of genetic modification using allelic replacement. This example shows a gene knockout being created in Pseudomonas aeruginosa. Here, sequences flanking the region to be deleted are cloned into the pEX18Tc vector, which contains a tetracycline resistance gene for selection of integrants and sacB, which permits selection with sucrose to promote the second recombination event and excision of the plasmid. Primer 1 and 2 are universal pEX18 forward and reverse primers that flank the multiple cloning sites, while primers 3 and 4 are specific to genomic DNA sequences flanking the sequences cloned into the pEX18Tc vector. Combinations of these are used to identify colonies in which integration and gene deletion have been successful. Credit: Huang W and Wilks A.4 Reproduced under the Creative Commons Attribution 4.0 International License.
Gene targeting revolutionized many genomics studies and its importance in advancing our understanding of mammalian biology was recognized in 2007 when the Nobel Prize was awarded to Dr. Mario R. Capecchi, Dr. Martin J. Evans and Dr. Oliver Smithies. However, creating mutant organisms using this approach can be time consuming and expensive. Thus, researchers continued to look for alternatives for which sequence-specific DNA endonucleases fitted the bill.
- Zinc finger nuclease (ZFN)
There are a whole range of restriction enzymes that will cut a wide variety of sequences, some with more specificity than others. However, most have short recognition sequences, meaning they will often cut many times in a genome. This is not necessarily a problem for generating inserts but is an issue when making precise cuts in the genome itself. However, Type II(S) restriction enzymes have separate DNA recognition and DNA cleavage domains, the cleavage domain of which will cut anything it is recruited to. Thus, by fusing the cleavage domain to a DNA binding domain that can be manipulated to target a sequence of choice, a precision genome cutting tool can be made. One such DNA binding domain is the zinc finger (ZF), the fusion of which is called a zinc finger nuclease (ZFN),5 developed in 19966 and first successfully used for genome mutagenesis in 20027 and modification in 2003.8
Each ZF binds 3 DNA bases, and by creating an array of ZF proteins (typically 3 or 4), a specific sequence (of 12 bases if 4 ZFs are used) can be recognized. The nuclease, typically FokI, will only cut as a dimer, meaning that a ZFN is required to bind either side of the target cut site, with 5–7 bases of space left in the middle, for nuclease activation. This helps to improve specificity and reduce off-target cleavage.
Mutations have also been made in some FokI nucleases making them obligate heterodimers, further reducing the likelihood of off-target activity. Repair may then occur via NHEJ, potentially introducing errors, which is useful for loss of function studies. By cutting twice, inversions and large deletions may be created. Insertions may also be made via NHEJ – e.g., inserting transgenes like a gene encoding a green florescent protein (GFP) reporter. However, you have no control over the orientation of the gene insertion and there will be indels flanking the insertion, causing the loss or gain of sequences which may be problematic depending on the purpose of the study. Depending on the mutations made, it may be possible to select for cells in which the desired modification has been made. If, on the other hand, a DNA template is present and the conditions favor HDR, a whole range of modifications including repairs and gene insertions can be made with accuracy.
ZFNs are quite small, making them easier than some alternatives to deliver into cells. However, recognition is typically limited to 24 bases (12 bases per side) as increasing the number of ZFs is generally unsuccessful. Assembling the ZF arrays is relatively simple; however, many ZFs don’t function well when assembled in arrays for reasons that aren’t fully understood, making self-assembly a laborious and often lengthy process with low success. Therefore, tried and tested commercial options tend to be preferable. Consequently, while they have their place and are a powerful genome editing tool, ZFNs have generally been superseded by alternative techniques, including transcription activator-like effector nucleases (TALENs), in recent years.
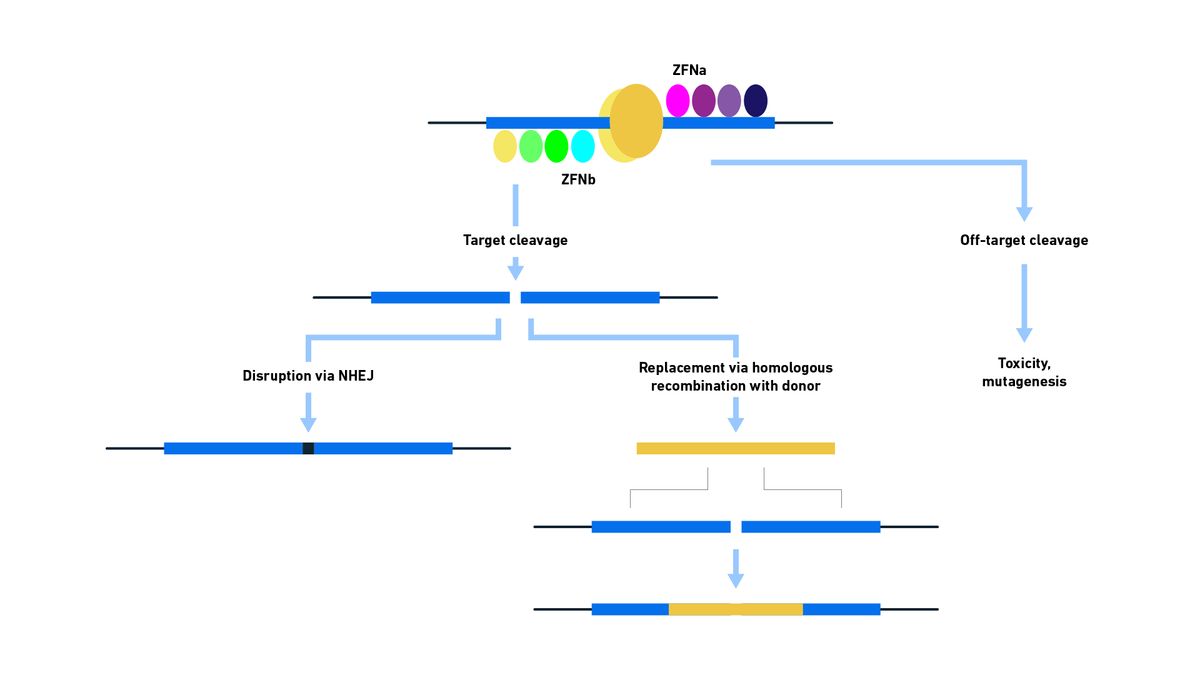
Figure 7: Schematic diagram showing the binding and activity of ZFNs used for genome modification. Following binding of the ZFN to the target sequence, cleavage occurs that may then be repaired by either NHEJ or HDR. Incorrect recognition of the target site may result in undesirable consequences. Credit: Technology Networks.
- Transcription activator-like effector nucleases (TALENs)
Developed in 2011,9 TALENs possess a DNA binding domain from a transcription activator-like effector (TALE) protein linked to a specific nuclease effector domain – an enzyme able to cut DNA.10 The TALE proteins contain repeat variable domains (RVDs), each one of which will bind to a specific base, unlike the triplets bound by ZFs (Figure 8). By experimentally manipulating the order of the RVDs into an array, TALEs can be made to recognize a specific DNA sequence, precisely targeting the TALEN to bind any desired target sequence in the genome.11 The associated nuclease, commonly FokI, is activated when dimerized, so TALENs have to be designed to bind either side of the position to be cut, very much like ZFNs, but with a larger gap of 14–20 bases in between. Once activated, the nuclease cuts the DNA in the middle of the gap between the bound TALE proteins, generating a four-base 5′ overhang, as with ZFNs. NHEJ will then remove this overhang during repair, making the process inherently mutagenic and introducing deletions, helpful for loss of gene function studies. If the repair occurs via HDR, various modifications, including insertions, deletions and inversions, can be introduced if homologous donor DNA is provided.
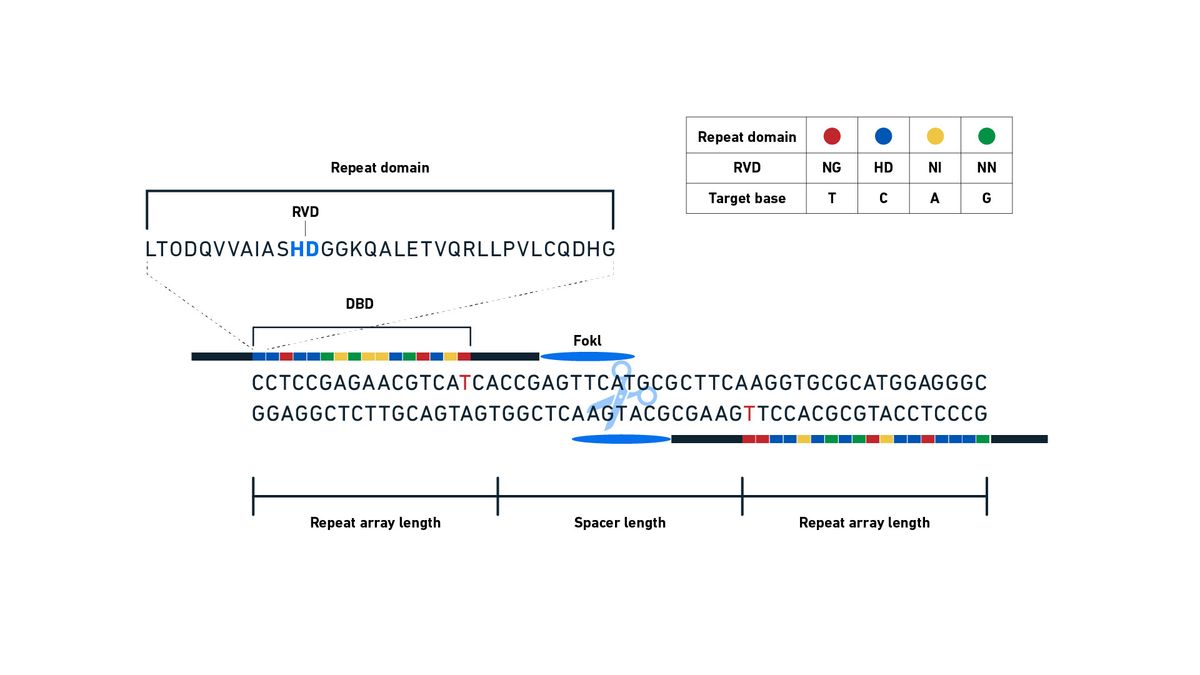
Figure 8: Schematic diagram showing TALEN structure, recognition sequences and cutting. Repeat domains each contain RVDs that can be arranged into an array to recognize a specific DNA sequence. When they bind successfully as a dimer, the FokI nuclease is activated and cuts the DNA, producing a DSB. Credit: Technology Networks.
TALENs are quite simple to assemble and have excellent specificity, offering recognition of 30–40 bases each, an improvement over ZFNs. They are particularly useful in hard-to-edit regions that clustered regularly interspaced short palindromic repeat (CRISPR) techniques struggle with or in regions lacking important CRISPR recognition sites. However, TALENs do require proteins (two per target) to be engineered prior to undertaking genome modifications, which can be expensive and time consuming, and making simultaneous edits can be challenging.
While in many cases, TALENs have been replaced by CRISPR technologies, they have been instrumental in many successes, including creating one of the earliest genome-edited crops, Xanthomonas-resistant rice,12 and the development of edited CAR-T cells used to eradicate tumors.13
- Homing endonucleases or meganucleases
Meganucleases, also called homing endonucleases, are naturally occurring site-specific nucleases with large recognition sites (typically 12–40 base pairs), helpful in precisely targeting a genomic site.14 They introduce DSBs at sites peculiar to the type of meganuclease, stimulating repair and offering the potential for modification of the cut site.
Consisting of five families, the recognition sites were limited by the naturally existing meganucleases available. Therefore, two approaches were taken to address this:15 either modifications were made to alter the specificity of existing examples, or chimeras capable of recognizing different sequences were created.
Successfully used in a number of applications, including gene therapy16 and crop engineering,17 meganucleases are considered less toxic to cells compared to some other methods like ZFNs. However, the construction of sequence-specific meganucleases is expensive and time-consuming compared to alternative approaches, making them a less popular choice.
- CRISPR gene editing
CRISPR–CRISPR-associated protein (Cas) systems (CRISPR–Cas systems) originate from RNA-based bacterial defense systems. They recognize and eliminate foreign DNA from invading plasmids and bacteriophage and are thus considered a type of bacterial “immune system”.
The system detects and cuts bacteriophage genomes, saving fragments of them (spacers) into repetitive CRISPR arrays as a record of previous infection so they can be targeted again in the future if they meet the same invader. This array is passed from generation to generation, growing with every new encounter (Figure 9).
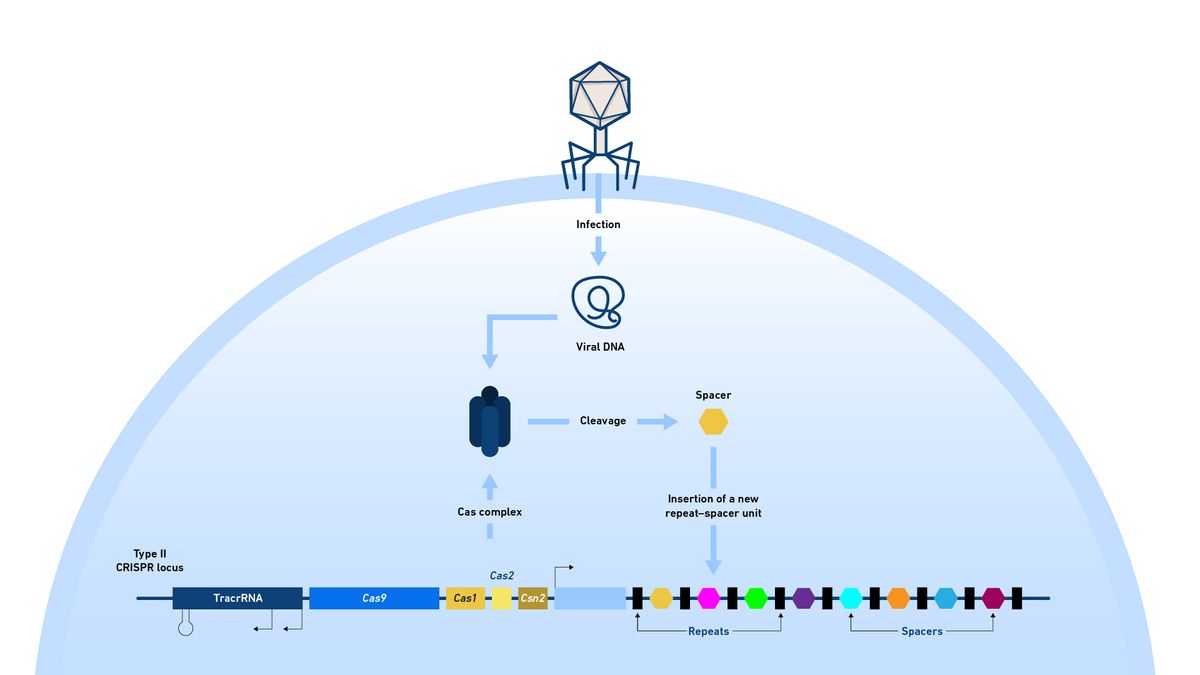
Figure 9: Diagram of a bacterial CRISPR locus indicating the genes included and CRISPR array. The route by which fragments of bacteriophage DNA may be added to the array is shown. The colored spacers represent all the fragments of bacteriophage genomes that the bacterium and its previous generations have encountered and cleaved. Credit: Technology Networks.
The CRISPR array of repeats and spacers is transcribed as a single pre-CRISPR transcript and the repeat regions bind the trans-activating CRISPR RNA (tracrRNA) in the locus. RNAse III then binds these double stranded sections and cleaves them, separating all the spacer and repeat pairs from the original single transcript, producing lots of CRISPR RNA–tracrRNA fragments in which the spacer sequence is now known as a protospacer (Figure 10). These fragments bind Cas9, an RNA guided DNA endonuclease, activating and programing it to search the genome for protospacer adjacent motifs (PAMs), which are often only three bases long. If it finds a PAM, Cas9 then unwinds the DNA and if there’s a match between the protospacer in its CRISPR RNA and the genomic DNA, Cas9 continues unwinding the DNA and cleaves the target.
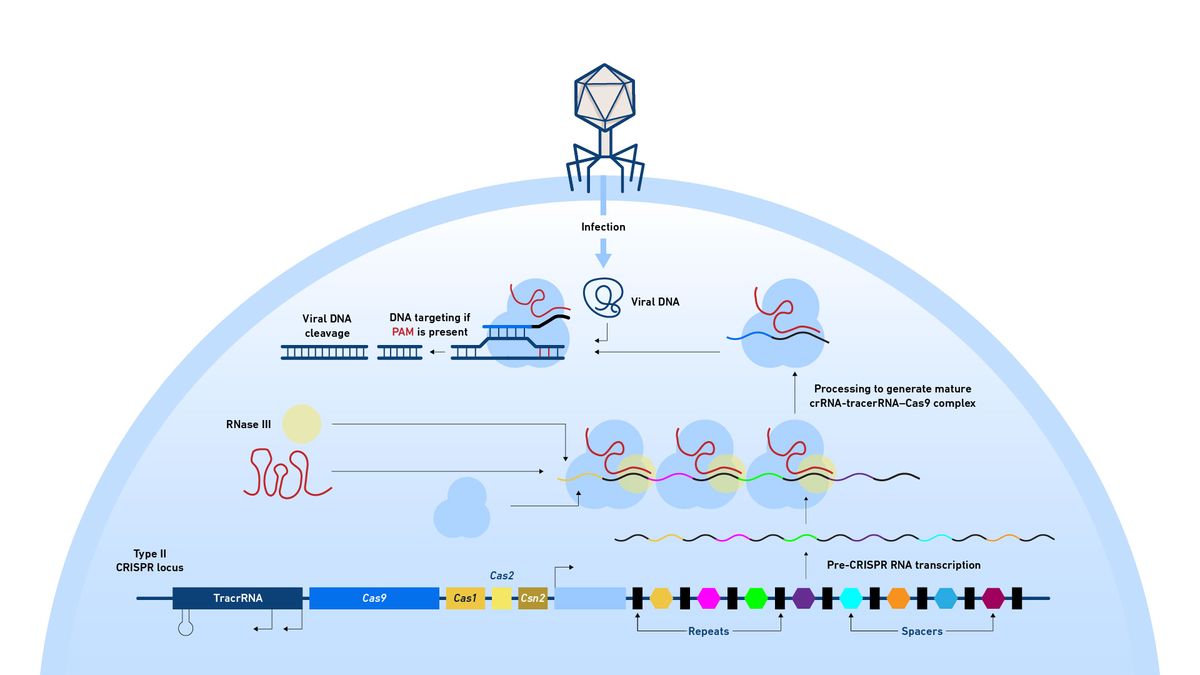
Figure 10: Diagram showing the mode of action of a CRISPR–Cas system. The process of locus replication, spacer cleavage and targeting of Cas9 to cleave DNA is indicated. Credit: Technology Networks.
Most Cas9 systems used in labs create a blunt-ended DSB three bases away from the PAM sequence, offering predictable cleavage. While the CRISPR–Cas system is a natural process, in 2012, Professors Emmanuelle Charpentier and Jennifer Doudna were able to show that it was a programable system,18 for which they received the Nobel Prize in 2020. Since then, scientists have been able to adapt it, using short guide RNAs (sgRNAs) to make DSBs at the very specific locations they require, and take advantage of NHEJ or HDR to repair the break and introduce mutations or make desired alterations (Figure 11).
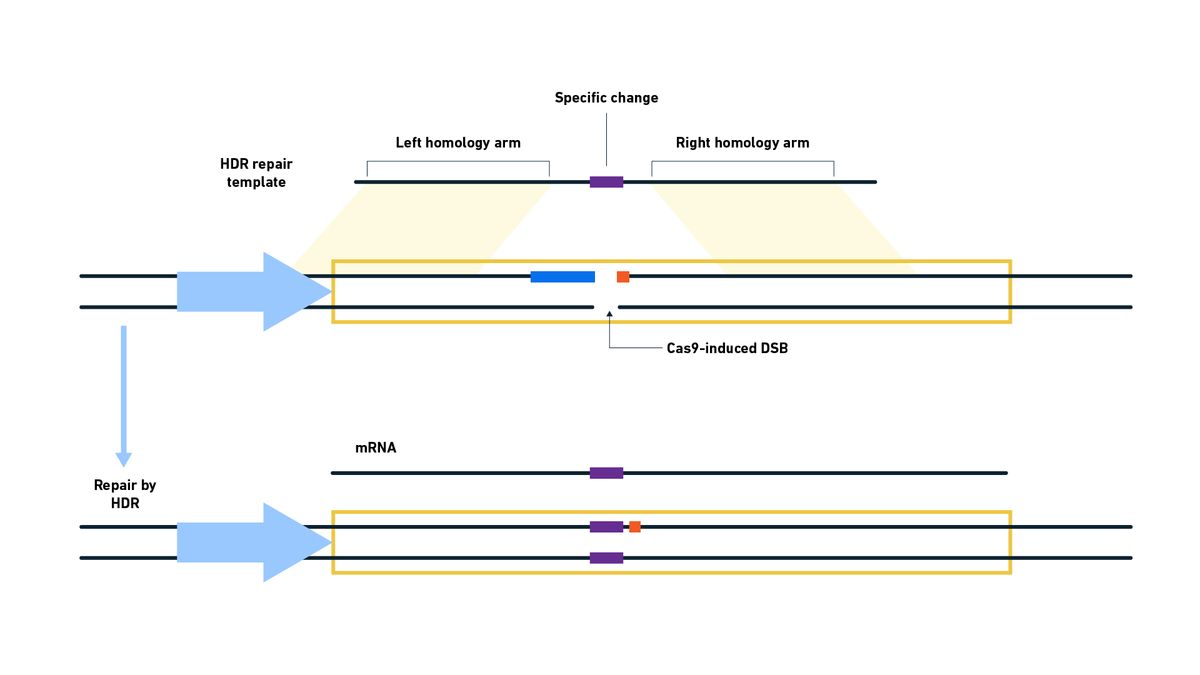
Figure 11: Following the DSB induced by Cas9, a template containing the desired alteration is used by the cell to repair the DSB via HDR, introducing the change into the genome. Credit: Technology Networks.
Cas9 can be programed to make a DSB wherever the user wishes in the genome. To do this, the Cas9 endonuclease is only supplied with a sgRNA that combines a tracr and protospacer sequence and is typically 98–100 bases long, 20 bases of which is the protospacer. Rather than being a phage derived sequence, the protospacer sequence is chosen by the scientist to target their desired genome location for cleavage (Figure 12).
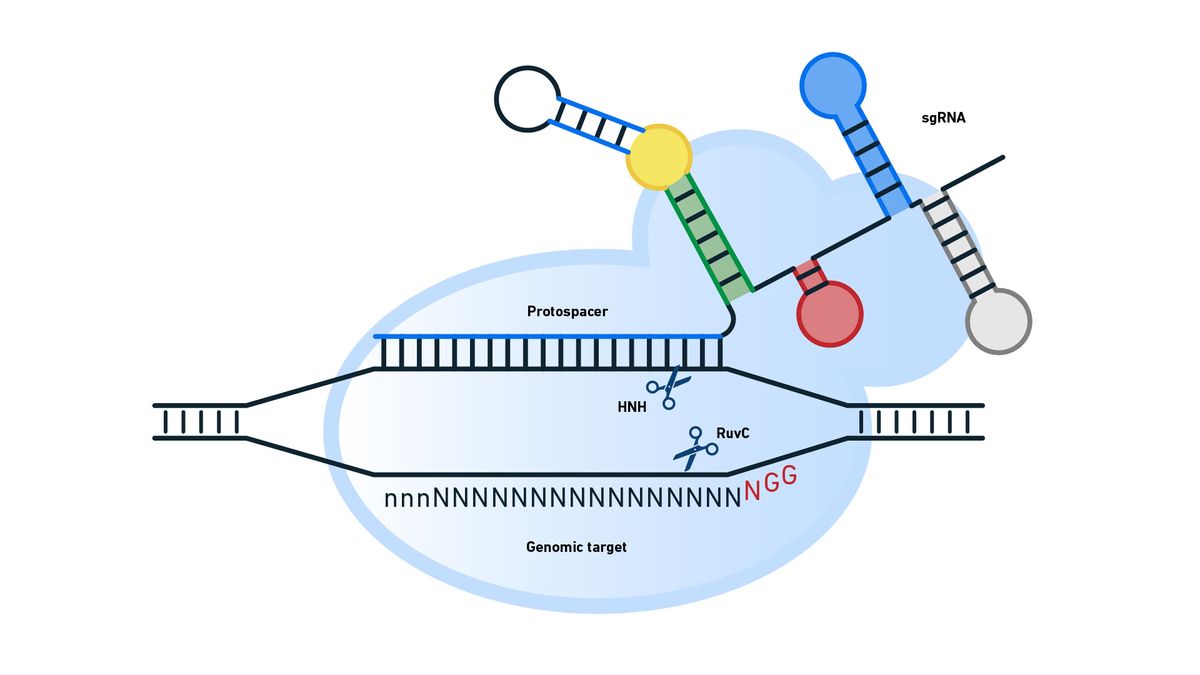
Figure 12: Cleavage of a desired genomic sequence by supplying Cas9 with a sgRNA. The enzymes involved in strand cleavage are indicated. Credit: Technology Networks.
Unlike TALENs, this system can easily be multiplexed, cutting many targets at once. The different guide RNAs just need to be delivered into cells at the same time as Cas9. These can be delivered to cells in a number of ways, including transfection of plasmids encoding Cas9 and the sgRNAs, transfection of Cas9 messenger RNA (mRNA) and synthesized sgRNA, transfection of recombinant Cas9 protein and synthesized sgRNAs and lentivirus transduction to deliver Cas9 and sgRNA expression vectors. Each has its own pros and cons, and the choice of which to use will depend on factors such as cost, time, cell type and whether you wish to target a single point or are performing a whole genome screen.
While CRISPR is very flexible, easy to use and good for multiplexing, it does make mistakes; this is useful in the setting of an adaptive immune system where bacteriophage may mutate to keep up with the arms race, but is not as helpful in genome modification. Cas9 will bind and sometimes cleave off-target sites that don’t perfectly match the protospacer, generating unwanted mutations. Scientists, however, have come up with a number of approaches to limit this activity including Cas9 mutants with enhanced specificity19 and dual nickase20 strategies.
Cas9 vs Cas3, Cas7-11, Cas 12a, Cas13 and Cas14
Cas9 is just one of the Cas family of proteins and probably the most studied. Other family members include Cas3, Cas7-11, Cas12a, Cas13 and Cas14, each with their own properties, cleavage methods and abilities. While Cas9 is popular due to its adaptability, utilizing other members of the Cas family of proteins for making genetic modifications may be desirable depending on your experimental goals. The properties of the different Cas family proteins are summarized in Table 1 below.
Table 1: Comparison of key features of the Cas family proteins and their uses for genetic modification.
|
| Cas3 | Cas9 | Cas7-11 | Cas12a | Cas13 | Cas14 |
| Best for targeting | DNA | DNA | Single stranded RNA (ssRNA) | DNA | ssRNA | Single stranded DNA (ssDNA) |
| Requirements for PAM or protospacer flanking sequence (PFS) | None | Various including NGG | None | TTN or TTTN | A, C or U | None |
| Notes | - Cuts by locating the target sequence and initiating ssDNA degradation unlike other Cas proteins - It can degrade several kilobases making it suited only to deletion generation | - Many variants exist offering different specialties and PAM sites - Breaks have blunt ends | - Functionally similar to Cas13 but less toxic to mammalian cells and doesn’t exhibit the non-specific RNA cleavage Cas13 does | - Breaks have sticky ends, good for HDR - Good for targeting AT-rich regions as the PAM doesn’t require any G’s or C’s - Smaller than Cas9 and doesn’t require a tracrRNA, making it easier to package for therapeutic delivery - Sensitive to mismatches in target sequence | - Can make transient edits as it targets RNA - Indiscriminately cleaves ssRNA when engaged at its RNA target sequence | - Sensitive to mismatched in the target sequence - Smaller than Cas9 - Indiscriminately cleaves non-complementary ssDNA when engaged by its target sequence |
| Applications | Making large deletions | Flexible for all types of mutant generation including insertions and deletions. Popular for knock outs | RNA editing and mRNA knockdown | Therapeutic delivery and HDR | RNA editing and mRNA knockdown. Some interest in its potential as a viral therapeutic | Shows promise for detecting rare, often hard-to-detect variants such as single nucleotide polymorphisms (SNPs) |
CRISPR are also divided into classes, primarily class 121 and class 2,22 based on their effector molecules and these are then further divided into subtypes.
Techniques for random modifications also exist, including:
- Mutagenesis induced by a mutagen
While not a technique that can be used to make precise genome modifications, mutagens such as nitrosoguanidine (NTG) can be used to introduce random point mutations throughout an organism’s genome. This approach has been used to investigate gene function and generate attenuated bacterial strains for vaccines,23 but its lack of controllability, mutational bias24 and potential for reversion25 means that it has largely been replaced by precision techniques.
- Random transgenesis
Using this technique, which has been popular in generating transgenic rodents26 and studying gene function27 due to protocol speed, exogenous DNA sequences or transposons are randomly integrated into the genome. The technique still finds utility among bacterial studies where pools of thousands of mutants can be generated through transposon insertion. These mutant pools can be tested in differing conditions and the impact on fitness of disrupting each gene can be assessed using next-generation sequencing (NGS) to compare the input and output pools, known as transposon directed insertion-site sequencing (TraDIS).28 However, the random nature of integration can produce unpredictable results for the target gene and surrounding genes, and all organisms generated require genotyping and phenotyping to understand a specific mutation. Therefore, as with mutagens, precision techniques are now often favored for producing individual mutants.
Gene editing pros and cons
There has been much debate over the safety and ethics of genetic modification. Some of the main positives and negatives that form the basis of these debates are summarized below in Table 2.
Table 2: Comparison of the main pros and cons debated with regard to genetic modification.
| Pros | Cons |
| Can prevent and/or treat genetic disease | May produce undesirable or unintended mutants with harmful properties or consequences |
| Can improve crop yield and pest resistance | May be open to abuse |
| Can produce or update vaccines in rapid response to a new or changing threat |
|
Applications of genome editing and genome engineering
The applications of genome editing and genetic engineering are numerous and diverse. Let’s consider some of the most important areas.
Crop engineering
With an ever-expanding population, there has been a growing need to maximize crop yields. This means not only cultivating crops that produce the maximum amount per plant, but that are also resistant to disease29 and tolerant of what may be unfavorable growth conditions30 to enable cultivation in inhospitable areas and in the face of our changing climate. This includes features such as tolerance to extremes of drought31 or flood,32 temperature33 and salt.34 Improved disease and pest resistance can also help to reduce the need for potentially expensive and/or harmful pesticide and chemical treatments.
Alterations in physical characteristics, such as plant height in wheat, can make the crops more robust and easier to harvest. Scientists have also been able to create crops with enhanced nutritional values, including rice35 and tomatoes,36 which is especially helpful in deprived areas where achieving a balanced diet can be challenging.
Flavor is another characteristic that has benefited from genetic modification. This has helped to counteract some declines in fruit and vegetable flavor that has occurred as a result of selective breeding for other traits37 such as size and firmness, for example in tomatoes.38 Researchers are also currently looking at removing flavor aspects that consumers dislike in foods such as brassicas.39
Despite widely publicized opposition to GM crops in the early days, over 190.4 million hectares of genetically engineered or biotech crops, spread across 29 countries, were planted in 2019.40
GM animals for food production
Much like the motivation for genetic modification of crops, changes to food animals can help to improve yield, disease resistance and flavor, remove allergens and reduce rearing times, thus production costs. Scientists are also using the approach to develop animals that generate less environmental pollution.
Examples include salmon that are genetically engineered to mature faster, 41 pigs lacking a sugar molecule known to trigger a rare allergy42 and cattle with a slicked hair type that could help to reduce temperature-related stress.43
Vaccine development
Genetic modification has been used for many years in the generation of vaccines,44 with the hepatitis B vaccine as an early example.45 Strains can be generated in which key genes have been mutated or removed to attenuate them and make them both safe and efficacious for use either alive (in live attenuated vaccines, such as the MMR vaccine46) or once killed (in inactivated vaccines, such as the polio vaccine47). Equally, the technique can be used to insert key genes associated with the generation of a protective immune response into a more generic backbone or carrier, as with viral vectored vaccines like those for Ebola and the ChAdOx1 SARS-CoV-2 vaccine.48 Alternatively, it can be used to make updates to existing vaccines to account for genetic change in the pathogen population, as is the case with the regular influenza vaccine updates.49 Even protein subunit vaccines, such as the meningitis B vaccine,50 rely on genetic modification in order to express the desired proteins in cells for purification.
Gene therapy
Gene therapy can be used to treat or prevent genetic diseases both inherited (such as sickle cell disease)51 and acquired (such as leukemia).52 While early techniques involved introducing new, non-faulty genes into cells, typically leaving the faulty gene in place, more recently, gene editing techniques have enabled scientists to instead correct these faults in the existing DNA. While gene therapy is widely seen as a positive step for health, the ethics of its use for germline editing53 to prevent disease in babies has divided people.54
Biomanufacturing
Genetic modification is a key technique for biomanufacturing and synthetic biology, providing the means with which to engineer a biological system to produce a desired gene product. This technique is used across many areas including the production of:
- Biopharmaceuticals such as insulin,55 growth hormones,56 human follicle-stimulating hormones (for treating infertility),57 human albumin,58 monoclonal antibodies,59 antihemophilic factors60 and vaccines.61
- Proteins for supplements and the future foods market.62
- Enzymes used in food production such as lipases, carbohydrases and proteases.63
As well as increasing production capacity and improving control over the systems, this approach allows scientists to make alterations to optimize the gene products for their application.
Creation of model animals and cell lines
Genetic modification techniques can be used to alter the genetics of model organisms or cell lines to make them more closely resemble the organism for which they serve as a model and consequently make them more representative. For example, mice are often used as a model to study human immunity; however, their immune system is different to ours in many ways. Consequently, genes can be altered, for example to express a given receptor of interest, to create “humanized” mice.64
The technique can also be used to make the specific mutations that occur in the human population and introduce them into model cell lines or organisms. This enables researchers to improve our understanding of the impacts of these changes, for example in neurodegenerative disease,65 and also provides a platform with which to investigate new vaccines and treatments.
Research into gene function
A prime way to find out what a gene does is to knock out its function and see what happens.66 By subsequently repairing the mutation or deletion that knocked it out, known as complementation,67 it is then possible to show that the change you made was responsible for the change seen and not as a result of some unintended off-target effect. The ability to make precise changes also means that not only the function of whole genes can be determined, but even certain parts of that gene and its gene product can too.
Adding fluorescent reporters
Inserting genes encoding fluorescent reporters allows cellular physical structures and processes to be monitored in real time,68 typically with the help of microscopy. The technique provides insights helpful in understanding dynamic regulation, spatial organization and the consequences of exposing cells or organisms to specific conditions or treatments. Fluorescent reporters may also help to reveal the consequences of other mutations or modifications that may have been made.
In conclusion, genetic modification techniques have evolved in their capabilities, usability and precision over the years. But whichever technique is used, it serves as an important tool for many applications, resulting in developments that we now take for granted in our daily lives, including life-saving and life-changing solutions.
1. Holden MTG, Heather Z, Paillot R, et al. Genomic evidence for the evolution of Streptococcus equi: Host restriction, increased virulence, and genetic exchange with human pathogens. PLoS Pathog. 2009;5(3):e1000346. doi:10.1371/journal.ppat.1000346
2. Mao Z, Bozzella M, Seluanov A, Gorbunova V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst). 2008;7(10):1765-1771. doi:10.1016/j.dnarep.2008.06.018
3. Scherer S, Davis RW. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci U S A. 1979;76(10):4951-4955. doi:10.1073/pnas.76.10.4951
4. Huang W, Wilks A. A rapid seamless method for gene knockout in Pseudomonas aeruginosa. BMC Microbiol. 2017;17(1):199. doi:10.1186/s12866-017-1112-5
5. Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636-646. doi:10.1038/nrg2842
6. Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci. 1996;93(3):1156-1160. doi:10.1073/pnas.93.3.1156
7. Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161(3):1169-1175. doi:10.1093/genetics/161.3.1169
8. Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300(5620):764-764. doi:10.1126/science.1079512
9. Cermak T, Doyle EL, Christian M, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39(12):e82. doi:10.1093/nar/gkr218
10. Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14(1):49-55. doi:10.1038/nrm3486
11. Becker S, Boch J. TALE and TALEN genome editing technologies. Gene Genome Ed. 2021;2:100007. doi:10.1016/j.ggedit.2021.100007
12. Li T, Liu B, Spalding MH, Weeks DP, Yang B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol. 2012;30(5):390-392. doi:10.1038/nbt.2199
13. Qasim W, Zhan H, Samarasinghe S, et al. Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Sci Transl Med. 2017;9(374):eaaj2013. doi:10.1126/scitranslmed.aaj2013
14. Silva G, Poirot L, Galetto R, et al. Meganucleases and other tools for targeted genome engineering: Perspectives and challenges for gene therapy. Curr Gene Ther. 2011;11(1):11-27. doi:10.2174/156652311794520111
15. Smith J, Grizot S, Arnould S, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34(22):e149. doi:10.1093/nar/gkl720
16. Chapdelaine P, Pichavant C, Rousseau J, Pâques F, Tremblay JP. Meganucleases can restore the reading frame of a mutated dystrophin. Gene Ther. 2010;17(7):846-858. doi:10.1038/gt.2010.26
17. D’Halluin K, Vanderstraeten C, Van Hulle J, et al. Targeted molecular trait stacking in cotton through targeted double-strand break induction. Plant Biotechnol J. 2013;11(8):933-941. doi:10.1111/pbi.12085
18. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816-821. doi:10.1126/science.1225829
19. Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351(6268):84-88. doi:10.1126/science.aad5227
20. Ran FA, Hsu PD, Lin CY, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380-1389. doi:10.1016/j.cell.2013.08.021
21. Makarova KS, Zhang F, Koonin EV. SnapShot: Class 1 CRISPR-Cas Systems. Cell. 2017;168(5):946-946.e1. doi:10.1016/j.cell.2017.02.018
22. Makarova KS, Zhang F, Koonin EV. SnapShot: Class 2 CRISPR-Cas Systems. Cell. 2017;168(1-2):328-328.e1. doi:10.1016/j.cell.2016.12.038
23. Cursons R, Patty O, Steward KF, Waller AS. Strangles in horses can be caused by vaccination with Pinnacle I. N. Vaccine. 2015;33(30):3440-3443. doi:10.1016/j.vaccine.2015.05.009
24. Harper M, Lee CJ. Genome-wide analysis of mutagenesis bias and context sensitivity of N-methyl-N′-nitro-N-nitrosoguanidine (NTG). Mutat. Res. - Fundam Mol. Mech. Mutagen. 2012;731(1):64-67. doi:10.1016/j.mrfmmm.2011.10.011
25. Klose SM, De Souza DP, Disint JF, et al. Reversion of mutations in a live mycoplasma vaccine alters its metabolism. Vaccine. 2023;41(21):3358-3366. doi:10.1016/j.vaccine.2023.04.045
26. Yan BW, Zhao YF, Cao WG, Li N, Gou KM. Mechanism of random integration of foreign DNA in transgenic mice. Transgenic Res. 2013;22(5):983-992. doi:10.1007/s11248-013-9701-z
27. Wudarski J, Ustyantsev K, Reinoite F, Berezikov E. Random integration transgenesis in a free-living regenerative flatworm Macrostomum lignano. In: Blanchoud S, Galliot B, eds. Whole-Body Regeneration: Methods and Protocols. Methods in Molecular Biology. Springer US; 2022:493-508. doi:10.1007/978-1-0716-2172-1_26
28. Langridge GC, Phan MD, Turner DJ, et al. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 2009;19(12):2308-2316. doi:10.1101/gr.097097.109
29. Gonsalves C, Lee DR, Gonsalves D. Transgenic virus-resistant papaya: The Hawaiian “rainbow” was rapidly adopted by farmers and is of major Importance in Hawaii today. apsnet.org. Published 2004. Accessed December 12, 2023. https://www.apsnet.org/edcenter/apsnetfeatures/Pages/PapayaHawaiianRainbow.aspx. doi:10.1094/APSnetFeature-2004-0804
30. Gajardo HA, Gómez-Espinoza O, Boscariol Ferreira P, Carrer H, Bravo LA. The potential of CRISPR/Cas technology to enhance crop performance on adverse soil conditions. Plants. 2023;12(9):1892. doi:10.3390/plants12091892
31. Sami A, Xue Z, Tazein S, et al. CRISPR–Cas9-based genetic engineering for crop improvement under drought stress. Bioengineered. 12(1):5814-5829. doi:10.1080/21655979.2021.1969831
32. White MD, Dalle Carbonare L, Lavilla Puerta M, et al. Structures of Arabidopsis thaliana oxygen-sensing plant cysteine oxidases 4 and 5 enable targeted manipulation of their activity. Proc Natl Acad Sci. 2020;117(37):23140-23147. doi:10.1073/pnas.2000206117
33. Singh A, Grover A. Genetic engineering for heat tolerance in plants. Physiol Mol Biol Plants. 2008;14(1-2):155-166. doi:10.1007/s12298-008-0014-2
34. Caine RS, Harrison EL, Sloan J, et al. The influences of stomatal size and density on rice abiotic stress resilience. New Phytol. 2023;237(6):2180-2195. doi:10.1111/nph.18704
35. Zafar S, Jianlong X. Recent advances to enhance nutritional quality of rice. Rice Sci. 2023;30(6):523-536. doi:10.1016/j.rsci.2023.05.004
36. Li J, Scarano A, Gonzalez NM, et al. Biofortified tomatoes provide a new route to vitamin D sufficiency. Nat Plants. 2022;8(6):611-616. doi:10.1038/s41477-022-01154-6
37. Shi Y, Vrebalov J, Zheng H, et al. A tomato LATERAL ORGAN BOUNDARIES transcription factor, SlLOB1, predominantly regulates cell wall and softening components of ripening. Proc Natl Acad Sci. 2021;118(33):e2102486118. doi:10.1073/pnas.2102486118
38. Davidovich-Rikanati R, Sitrit Y, Tadmor Y, et al. Enrichment of tomato flavor by diversion of the early plastidial terpenoid pathway. Nat Biotechnol. 2007;25(8):899-901. doi:10.1038/nbt1312
39. Brown B. The first CRISPR-edited salad is here. pairwise.com. Published May 16, 2023. Accessed December 12, 2023. https://www.pairwise.com/news/the-first-crispr-edited-salad-is-here
40. ISAAA. ISAAA Brief 55-2019: Executive Summary. ISAAA.org. Accessed December 12, 2023. https://www.isaaa.org/resources/publications/briefs/55/executivesummary/default.asp
41. Bodnar A. Fast-growing genetically engineered salmon approved. Biology Fortified Inc. Published March 12, 2019. Accessed December 12, 2023. http://biofortified.org/2019/03/gmo-salmon-approved/
42. Cima G. FDA approves genetic alteration in pigs. American Veterinary Medical Association avma.org. Published January 21, 2021. Accessed December 12, 2023. https://www.avma.org/javma-news/2021-02-01/fda-approves-genetic-alteration-pigs
43. Cima G. Genetically modified cattle may be sold for food in U.S. American Veterinary Medical Association avma.org. Published March 21, 2022. Accessed December 12, 2023. https://www.avma.org/news/genetically-modified-cattle-may-be-sold-food-us
44. Murray K, Stahl S, Ashton-Rickardt PG, Potter G. Genetic engineering applied to the development of vaccines. Philosoph. Trans. R. Soc. B, Bio. Sci. 1997;324(1224):461-476. doi:10.1098/rstb.1989.0060
45. Genetically engineered hepatitis B vaccine now available. CMAJ. 1987;137(4):301. PMCID:PMC1494025
46. World Health Organization. Measles, mumps and rubella (MMR). who.int. Accessed December 12, 2023. https://www.who.int/teams/health-product-policy-and-standards/standards-and-specifications/vaccines-quality/measles-mumps-and-rubella-(mmr)
47. World Health Organization. History of polio vaccination. who.int. Accessed December 12, 2023. https://www.who.int/news-room/spotlight/history-of-vaccination/history-of-polio-vaccination
48. Folegatti PM, Ewer KJ, Aley PK, et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: A preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396(10249):467-478. doi:10.1016/S0140-6736(20)31604-4
49. Centers for Disease Control and Prevention. Selecting viruses for the seasonal flu vaccine. cdc.gov. Published November 3, 2022. Accessed December 12, 2023. https://www.cdc.gov/flu/prevent/vaccine-selection.htm
50. Bianchi A, Fantoni S, Prugnola A. Meningococcal B vaccine and the vision of a meningitis free world. J Prev Med Hyg. 2015;56(3):E140-E143. PMCID:PMC4755123
51. UK Government. MHRA authorises world-first gene therapy that aims to cure sickle-cell disease and transfusion-dependent β-thalassemia. GOV.UK. Accessed December 12, 2023. https://www.gov.uk/government/news/mhra-authorises-world-first-gene-therapy-that-aims-to-cure-sickle-cell-disease-and-transfusion-dependent-thalassemia
52. Reardon S. Leukaemia success heralds wave of gene-editing therapies. Nature. 2015;527(7577):146-147. doi:10.1038/nature.2015.18737
53. Wolf DP, Mitalipov PA, Mitalipov SM. Principles of and strategies for germline gene therapy. Nat Med. 2019;25(6):890-897. doi:10.1038/s41591-019-0473-8
54. Rainie L, Funk C, Anderson M Tyson A. Americans are closely divided over editing a baby’s genes to reduce serious health risk. Pew Research Center: Internet, Science & Tech. pewresearch.org. Published March 17, 2022. Accessed December 12, 2023. https://www.pewresearch.org/internet/2022/03/17/americans-are-closely-divided-over-editing-a-babys-genes-to-reduce-serious-health-risk/
55. Baeshen NA, Baeshen MN, Sheikh A, et al. Cell factories for insulin production. Microb Cell Fact. 2014;13:141. doi:10.1186/s12934-014-0141-0
56. Blethen SL, Baptista J, Kuntze J, Foley T, LaFranchi S, Johanson A. Adult height in growth hormone (GH)-deficient children treated with biosynthetic GH. J Clin Endocrinol Metab. 1997;82(2):418-420. doi:10.1210/jcem.82.2.3734
57. Winstel R, Wieland J, Gertz B, Mueller A, Allgaier H. Manufacturing of recombinant human follicle-stimulating hormone Ovaleap® (XM17), comparability with Gonal-f®, and performance/consistency. Drugs R D. 2017;17(2):305. doi:10.1007/s40268-017-0182-z
58. Chuang VTG, Otagiri M. Recombinant human serum albumin. Drugs Today (Barc). 2007;43(8):547-561. doi:10.1358/dot.2007.43.8.1067343
59. DeLuca KF, Mick JE, DeLuca JG. Production and purification of recombinant monoclonal antibodies from human cells based on a primary sequence. STAR Protoc. 2022;3(4):101915. doi:10.1016/j.xpro.2022.101915
60. Antihemophilic factor [recombinant], formulated with sucrose. P T. 2010;35(9 Section 3):2-5. PMCID:PMC2957727
61. Types of vaccine. ox.ac.uk. Accessed December 12, 2023. https://vaccineknowledge.ox.ac.uk/types-of-vaccine
62. Hettinga K, Bijl E. Can recombinant milk proteins replace those produced by animals? Curr Opin Biotechnol. 2022;75:102690. doi:10.1016/j.copbio.2022.102690
63. Zhang Y, Geary T, Simpson BK. Genetically modified food enzymes: a review. Curr Opin Food Sci. 2019;25:14-18. doi:10.1016/j.cofs.2019.01.002
64. Pearson T, Greiner DL, Shultz LD. Creation of “humanized” mice to study human immunity. Curr Protoc Immunol. 2008; 81:15.21.1-15.21.21. doi:10.1002/0471142735.im1521s81
65. Yang W, Chen X, Li S, Li XJ. Genetically modified large animal models for investigating neurodegenerative diseases. Cell Biosci. 2021;11:218. doi:10.1186/s13578-021-00729-8
66. Housden BE, Muhar M, Gemberling M, et al. Loss-of-function genetic tools for animal models: cross-species and cross-platform differences. Nat Rev Genet. 2017;18(1):24-40. doi:10.1038/nrg.2016.118
67. Kumar D. Synthetic Gene complementation to determine off-target silencing. In: Mysore KS, Senthil-Kumar M, eds. Plant Gene Silencing: Methods and Protocols. Methods in Molecular Biology. Springer; 2015:281-293. doi:10.1007/978-1-4939-2453-0_21
68. Mehta S, Zhang J. Reporting from the field: Genetically encoded fluorescent reporters uncover signaling dynamics in living biological systems. Annu Rev Biochem. 2011;80:375-401. doi:10.1146/annurev-biochem-060409-093259